Figure 2.
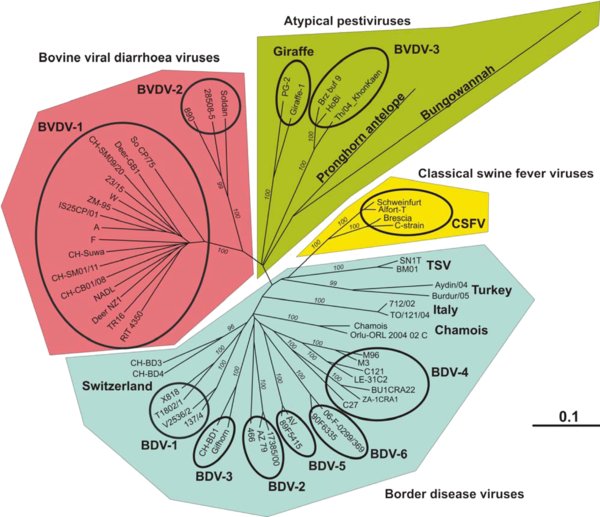
Phylogenetic analysis and classification of pestiviruses based on Npro: The phylogenetic tree that represents the genetic relationship of pestivirus strains was constructed from the complete Npro coding sequences except for the two strains Aydin/04 (EU930014) and Burdur/05 (EU930015). However, since constructing the tree with all sequences shortened to 420 nucleotides did not change the basic structure of the tree (data not shown), we included these strains in the analysis. Sequences were obtained from GenBank and the sequences of the Swiss BDV strains CH-BD3 (GU244490) and CH-BD4 (GU244489) were described by C. Reichert1. The analysis was computed using the programs included in the GCG software package as PILEUP for the multiple sequence alignment and PAUP for calculating the distances by the Kimura 2-parameter method and constructing the tree according to the neighbor-joining method. Branch numbers (italics) indicate the percentage of 1 000 bootstrap replicates. Only bootstrap values over 95 are shown. Branch lengths are proportional to genetic distances, and the bar shown indicates 0.1 substitutions per site. The classification of the various pestiviruses was composed according to [23, 71, 94]. Virus strains within the “BVDV-3” group were named according to [56, 57], but they had previously also been labeled as “unclassified” [90] or “atypical” [55]. Virus strains within the chamois group in this paper were sometimes assigned to BDV-4 [3, 61, 71, 78, 93, 99], whereas the Tunisian isolates SN1T and BM01 were recently labeled as TSV (Tunisian sheep virus, [57]). (A color version of this figure is available at www.vetres.org.)


